Integration Of Python And Keras In Anaconda In To Knime For Mac
Allows execution of a Python script in a local Python installation. The path to the Python executable has to be configured in Preferences → KNIME → Python. This node supports Python 2 and 3. It also allows to import Jupyter notebooks as Python modules via the knimejupyter module that is part of the Python workspace.
In my previous I wrote about my first experiences with KNIME and we implemented three classical supervised machine learning models to detect credit card fraud. In the meantime I found out that the newest version of KNIME (at this time 3.6) supports also the deep learning frameworks TensorFlow and Keras. So I thought lets revisit our deep learning model for theand try to implement in KNIME using Keras without writing one line of Python code.
Install the required packagesThe requirement is you have Python with TensorFlow and Keras (you can install it with pip or conda, if you using the anaconda distribution) on your machine. Then you need to install the Keras integration extensions in KNIME, you can follow the official tutorial on. At the moment I am quite busy with preparing a two training course about ‘Programming and Quantitative Finance in Python’ and ‘Programming and Machine Learning in Python’ for internal trainings at my work, so I haven’t had much free time for my blog. But I spent the last two nights fooling around with, an open-source tool for data analytics / mining (Peter, I took inspiration from your to name today’s post) and I want to share my experience. In the beginning I was quite sceptical and my first thought was ‘I can write code faster then drag-n-drop a model’ (and I still believe it). But I wanted to give it a try and I migrated my logistic regression fraud detection sample from my previous blog into a graphical workflow.In the beginning it was bit frustrating since I didn’t know which node to use and where to find all the settings.
But the interface and the node names are quite self-explaining so after exploring some examples and watching one or two youtube videos I was able build my first fraud detection model in KNIME.To work with a classifier we need to transfer the numerical variable Class (1=Fraud, 0=NonFraud) into a string variable. This was not obvious for me and coming from Python and SkLearn it felt a bit wired and unnecessary.
After fixing that, I split the data in a training and test set with the Partioning node. Deezer hifi. With a right-click on the node we can adjust the configuration and can change the it to the common 80-20 split.In the next step the data will be standardized (using a normalizer node). We can select which features / variable and how we want to scale them. We have plenty of settings to choose from, a nice feature is the online help (node description) on the right side of the UI, which describes the different parameter.I have capsuled the model fitting, prediction and scoring into a meta-node to make the workflow look cleaner and more understandable. With a double-click on the meta-node we can open the sub-workflow.I am fitting three different models (again encapsulated into meta-nodes) and combine the results (the AUC Score) into one table and write it and export it as a csv-file.Lets have a detailed look into the logistic regression model meta-node.The first node is the so-called learner node. It fits the model on the training data.
In the configuration we can select the input features, target column, the solver algorithm, and advanced settings like regularizations.To make predictions we use the predictor node. The inputs are the fitted model (square input) and the standardized test set. Into the normaliser (apply) node, we feed the test set and the fitted normalizer (as far as I understand that is equivalent to use the transform method of a Scaler in Sklearn after fitting it before, please correct me if I am wrong). The prediction output will be used to calculate the AUC curve (in the configuration setting of the prediction node we have to add the predicted probabilities as an additional output, in the default settings is to output only the predicted class). We export the plot as an SVG file and the auc score (as a table with an extra column for the model name) is the output of our meta-node.We can always investigate the output/result of one step, e.g.
Of the last node:Or the interactive plot of the AUC node:Or the model parameter output of the learner node:The workflow for the other two models is quite similar. With copy and pasting the Logistic Regression meta-node, it was just replacing the learner and predictor node and adjusting the configurations.To execute the complete workflow we just need to press the run/play button in the menu.There is still much to discover and explore and try. For example there are node for cross-validation and feature selection which I haven’t tried yet and so many other nodes (e.g plotting, descriptive statistics and the Python and R integration nodes). And I haven’t tried to move a model into production, but I read that it should not be that difficult with KNIME (they promote it as a platform to create data science applications). I spent just a couple hours with it, so please forgive me if I didn’t use the right name for some of the nodes, setting, menus or features in KNIME.What is my impression after playing with it for a couple hours?I still believe that writing code is the faster option for me, but I have to admit that I like it more and more. And its not really fair comparison (years of Python programming vs couple hours experimenting with a new tool). Its a nice tool for prototyping models without writing a line of code. If you are not familiar with a ML library yet, its a good and fast way to build models.
But here is no free lunch either, instead of learning a syntax and the architecture of a library you have to learn to use the UI and find all the settings.It’s an open-source software and so far I haven’t encountered any limitation (e.g. Other tools limit the numbers of rows you can use in a free version) but I’ve just scratched the surface.In my opinion one big advantage is the visualization of the model. Eforcity 9 to 4 pin ieee 1394b firewire 800 cable for mac os. The model is easy to understand and can easily be handed over to some other developers or engineer. Everyone knows that working with other people’s code can be a sometimes a pain and having a visual workflow can eliminate that pain. But I believe the workflows can become messy as well.
Its a tool which can be used by analysts and business user who want to explore and analyse their data, generate insights and use the power of standard machine learning and data mining algorithm without being forced to learn programming first.The first impression is surprisingly good and I will continue playing with it and I want to figure out how to run my own Python script in a node and maybe even more important how to move a model into production.I will report about it in a later post.So long.
In my previous blog post, I showed how to use the KNIME Deep Learning/DL4J Integration to predict the handwritten digits from images in the MNIST dataset. That's a neat trick, but it's a problem that has been pretty well-solved for a while. What about trying something a bit more difficult? In this post, I'll take a dataset of images from three different subtypes of lymphoma and classify the image into the (hopefully) correct subtype.brings new deep learning capabilities to KNIME Analytics Platform. You can now use the Keras Python library to take advantage of a variety of different deep learning backends. The new KNIME nodes provide a convenient GUI for training and deploying deep learning models while still allowing model creation/editing directly in Python for maximum flexibility.The workflows mentioned in this post require a fairly heavy amount of computation (and waiting), so if you're just looking to check out the new integration, here that recapitulates the results of the previous blog post using the new Keras integration.
There are quite a few more example workflows for both DL4J and Keras, which can be found in.Right, back to the challenge. Malignant lymphoma affects many people, and among malignant lymphomas, CLL (chronic lymphocytic leukemia), FL (follicular lymphoma), and MCL (mantle cell lymphoma) are difficult for even experienced pathologists to accurately classify.A typical task for a pathologist in a hospital would be to look at those images and make a decision about what type of lymphoma is present. In many cases, follow-up tests to confirm the diagnosis are required.
An assistive technology that can guide the pathologist and speed up their job would be of great value. Freeing up the pathologist to spend their time on those tasks that computers can't do so well, has obvious benefits for the hospital, the pathologist, and the patients.Figure 1: The modeling process adopted to classify lymphoma images. At each stage, the required components are listed.
Getting the DatasetSince I just have my laptop and Keras runs really fast on modern GPUs, I'll be logging into KNIME Analytics Platform hosted on an Azure N-series GPU. You could, of course, do the same using an AWS equivalent or a fast GPU in your workstation. Full details on.The full dataset is available as. I created a workflow that downloads the file and extracts the images into three sub-directories — one for each lymphoma type. Finally, I created a KNIME table file that stores the path to the image files and labels the image according to the class of lymphoma. Preparing the ImagesEach image has the dimensions 1388 by 1040px and the information required to determine the classification is a property of the whole image (i.e.
It's not reliant only on individual cells, which can be the case in some image classification problems). In the next step, we'll use the to train a classifier.
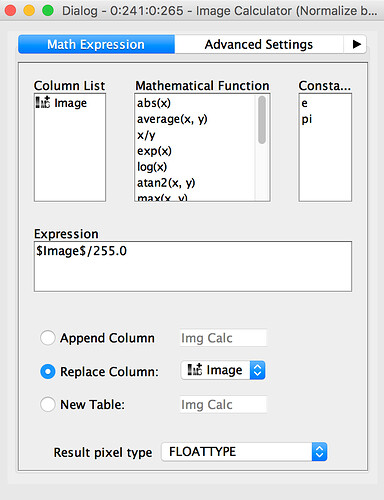
This model expects image sizes of 64 by 64px, so we'll need to chop the whole images into multiple patches that we'll then use for learning.Figure 2: Workflow to load and preprocess the image files. The results are small image patches of 64x64px.The general workflow just splits the input KNIME table into two datasets (train and test). Inside the Load and preprocess images (Local Files) wrapped metanode, we use the KNIME Image Processing extension to read the image file, normalize the full image, and then crop and split the image into 64 by 64px patches. Once again, we write out a KNIME table file, this time containing the image patches and the class information.Figure 3: The steps to cut a full image into multiple 64x64px image patches, contained in wrapped metanodes named Load and preprocess images (Local Files). Training the ModelSince developing a completely novel CNN is both difficult and time-consuming, we're going to first try using an existing CNN that has been pre-trained for solving image classification problems.